
|
Convolution Filters |  |
Modules | |
| Parallel Processing | |
Functions | |
| template<... > | |
| void | convolveFFT (...) |
| Convolve an array with a kernel by means of the Fourier transform. More... | |
| template<... > | |
| void | convolveFFTComplex (...) |
| Convolve a complex-valued array by means of the Fourier transform. More... | |
| template<... > | |
| void | convolveFFTComplexMany (...) |
| Convolve a complex-valued array with a sequence of kernels by means of the Fourier transform. More... | |
| template<... > | |
| void | convolveFFTMany (...) |
| Convolve a real-valued array with a sequence of kernels by means of the Fourier transform. More... | |
| template<... > | |
| void | convolveImage (...) |
| Convolve an image with the given kernel(s). More... | |
| template<... > | |
| void | convolveImageWithMask (...) |
| Deprecated name of 2-dimensional normalized convolution, i.e. convolution with a mask image. More... | |
| template<... > | |
| void | convolveMultiArrayOneDimension (...) |
| Convolution along a single dimension of a multi-dimensional arrays. More... | |
| template<... > | |
| void | correlateFFT (...) |
| Correlate an array with a kernel by means of the Fourier transform. More... | |
| template<... > | |
| void | gaussianDivergenceMultiArray (...) |
| Calculate the divergence of a vector field using Gaussian derivative filters. More... | |
| template<... > | |
| void | gaussianGradient (...) |
| Calculate the gradient vector by means of a 1st derivatives of Gaussian filter. More... | |
| template<... > | |
| void | gaussianGradientMagnitude (...) |
| Calculate the gradient magnitude by means of a 1st derivatives of Gaussian filter. More... | |
| template<... > | |
| void | gaussianGradientMultiArray (...) |
| Calculate Gaussian gradient of a multi-dimensional arrays. More... | |
| template<... > | |
| void | gaussianSharpening (...) |
| Perform sharpening function with gaussian filter. More... | |
| template<... > | |
| void | gaussianSmoothing (...) |
| Perform isotropic Gaussian convolution. More... | |
| template<... > | |
| void | gaussianSmoothMultiArray (...) |
| Isotropic Gaussian smoothing of a multi-dimensional arrays. More... | |
| template<... > | |
| void | hessianMatrixOfGaussian (...) |
| Filter image with the 2nd derivatives of the Gaussian at the given scale to get the Hessian matrix. More... | |
| template<... > | |
| void | hessianOfGaussianMultiArray (...) |
| Calculate Hessian matrix of a N-dimensional arrays using Gaussian derivative filters. More... | |
| template<... > | |
| void | laplacianOfGaussian (...) |
| Filter image with the Laplacian of Gaussian operator at the given scale. More... | |
| template<... > | |
| void | laplacianOfGaussianMultiArray (...) |
| Calculate Laplacian of a N-dimensional arrays using Gaussian derivative filters. More... | |
| template<... > | |
| void | normalizedConvolveImage (...) |
| Performs a 2-dimensional normalized convolution, i.e. convolution with a mask image. More... | |
| template<... > | |
| void | rieszTransformOfLOG (...) |
| Calculate Riesz transforms of the Laplacian of Gaussian. More... | |
| template<... > | |
| void | separableConvolveMultiArray (...) |
| Separated convolution on multi-dimensional arrays. More... | |
| template<... > | |
| void | simpleSharpening (...) |
| Perform simple sharpening function. More... | |
| template<... > | |
| void | structureTensor (...) |
| Calculate the Structure Tensor for each pixel of and image, using Gaussian (derivative) filters. More... | |
| template<... > | |
| void | structureTensorMultiArray (...) |
| Calculate the structure tensor of a multi-dimensional arrays. More... | |
| template<... > | |
| void | symmetricGradientMultiArray (...) |
| Calculate gradient of a multi-dimensional arrays using symmetric difference filters. More... | |
Detailed Description
The functions in this group implement separable convolutions (e.g. smoothing and sharpening, Gaussian derivatives) and related filters (like the gradient magnitude) on arbitrary-dimensional arrays. In addition, non-separable filters are supported for 2D images. All functions accept the vigra::MultiArrayView API (which can be wrapped around a wide variety of data structures) as well as a number of deprecated APIs such as Image Iterators.
Function Documentation
| void vigra::rieszTransformOfLOG | ( | ... | ) |
Calculate Riesz transforms of the Laplacian of Gaussian.
The Riesz transforms of the Laplacian of Gaussian have the following transfer functions (defined in a polar coordinate representation of the frequency domain):
![\[ F_{\sigma}(r, \phi)=(i \cos \phi)^n (i \sin \phi)^m r^2 e^{-r^2 \sigma^2 / 2} \]](form_4.png)
where n = xorder and m = yorder determine th e order of the transform, and sigma > 0 is the scale of the Laplacian of Gaussian. This function computes a good spatial domain approximation of these transforms for xorder + yorder <= 2. The filter responses may be used to calculate the monogenic signal or the boundary tensor.
Declarations:
pass 2D array views:
Usage:
#include <vigra/boundarytensor.hxx>
Namespace: vigra
| void vigra::convolveImage | ( | ... | ) |
Convolve an image with the given kernel(s).
If you pass vigra::Kernel2D to this function, it will perform an explicit 2-dimensional convolution. If you pass a single vigra::Kernel1D, it performs a separable convolution, i.e. it concatenates two 1D convolutions (along the x-axis and along the y-axis) with the same kernel via internal calls to separableConvolveX() and separableConvolveY(). If two 1D kernels are specified, separable convolution uses different kernels for the x- and y-axis.
All border treatment modes are supported.
The input pixel type T1 must be a linear space over the kernel's value_type T, i.e. addition of source values, multiplication with kernel values, and NumericTraits must be defined. The kernel's value_type must be an algebraic field, i.e. the arithmetic operations (+, -, *, /) and NumericTraits must be defined. Typically, you will use double for the kernel type.
Declarations:
pass 2D array views:
Usage:
#include <vigra/convolution.hxx>
Namespace: vigra
Preconditions:
The image must be larger than the kernel radius.
-
For 1D kernels,
w > std::max(xkernel.right(), -xkernel.keft())andh > std::max(ykernel.right(), -ykernel.left())are required. -
For 2D kernels,
w > std::max(kernel.lowerRight().x, -kernel.upperLeft().x)andh > std::max(kernel.lowerRight().y, -kernel.upperLeft().y)are required.
If BORDER_TREATMENT_CLIP is requested: the sum of kernel elements must be != 0.
- Examples:
- smooth_convolve.cxx.
| void vigra::simpleSharpening | ( | ... | ) |
Perform simple sharpening function.
This function uses convolveImage() with the following 3x3 filter:
and uses BORDER_TREATMENT_REFLECT as border treatment mode.
Preconditions:
Declarations:
pass 2D array views:
Usage:
#include <vigra/convolution.hxx>
Namespace: vigra
| void vigra::gaussianSharpening | ( | ... | ) |
Perform sharpening function with gaussian filter.
This function uses gaussianSmoothing() at the given scale to create a temporary image 'smooth' and than blends the original and smoothed image according to the formula
Preconditions:
Declarations:
pass 2D array views:
Usage:
#include <vigra/convolution.hxx>
Namespace: vigra
| void vigra::gaussianSmoothing | ( | ... | ) |
Perform isotropic Gaussian convolution.
This function is a shorthand for the concatenation of a call to separableConvolveX() and separableConvolveY() with a Gaussian kernel of the given scale. If two scales are provided, smoothing in x and y direction will have different strength. The function uses BORDER_TREATMENT_REFLECT.
Function gaussianSmoothMultiArray() performs the same filter operation on arbitrary dimensional arrays.
Declarations:
pass 2D array views:
Usage:
#include <vigra/convolution.hxx>
Namespace: vigra
- Examples:
- smooth.cxx.
| void vigra::gaussianGradient | ( | ... | ) |
Calculate the gradient vector by means of a 1st derivatives of Gaussian filter.
This function is a shorthand for the concatenation of a call to separableConvolveX() and separableConvolveY() with the appropriate kernels at the given scale. Note that this function can either produce two separate result images for the x- and y-components of the gradient, or write into a vector valued image (with at least two components).
Function gaussianGradientMultiArray() performs the same filter operation on arbitrary dimensional arrays.
Declarations:
pass 2D array views:
Usage:
#include <vigra/convolution.hxx>
Namespace: vigra
| void vigra::gaussianGradientMagnitude | ( | ... | ) |
Calculate the gradient magnitude by means of a 1st derivatives of Gaussian filter.
This function calls gaussianGradient() and returns the pixel-wise magnitude of the resulting gradient vectors. If the original image has multiple bands, the squared gradient magnitude is computed for each band separately, and the return value is the square root of the sum of these squared magnitudes.
Anisotropic data should be provided with appropriate vigra::ConvolutionOptions to adjust the filter sizes for the resolution of each axis. Otherwise, the parameter opt is optional unless the parameter sigma is omitted.
If you pass vigra::BlockwiseConvolutionOptions instead, the algorithm will be executed in parallel on data blocks of a certain size. The block size can be customized via BlockwiseConvolutionOptions::blockShape(), but the defaults usually work reasonably. By default, the number of threads equals the capabilities of your hardware, but you can change this via BlockwiseConvolutionOptions::numThreads().
Declarations:
use arbitrary-dimensional arrays:
Here, the input element types T1 and MT can be arbitrary scalar types, and T1 may also be TinyVector or RGBValue. The output element type T2 should be the corresponding norm type (see NormTraits). In the Multiband<MT>-version, the input array's right-most dimension is interpreted as a channel axis, therefore it must have one dimension more than the output array.
Usage:
#include <vigra/multi_convolution.hxx> (sequential version)
#include <vigra/multi_blockwise.hxx> (parallel version)
#include <vigra/convolution.hxx> (deprecated API version)
Namespace: vigra
- Examples:
- graph_agglomerative_clustering.cxx, and watershed.cxx.
| void vigra::laplacianOfGaussian | ( | ... | ) |
Filter image with the Laplacian of Gaussian operator at the given scale.
This function calls separableConvolveX() and separableConvolveY() with the appropriate 2nd derivative of Gaussian kernels in x- and y-direction and then sums the results to get the Laplacian.
Function laplacianOfGaussianMultiArray() performs the same filter operation on arbitrary dimensional arrays.
Declarations:
pass 2D array views:
Usage:
#include <vigra/convolution.hxx>
Namespace: vigra
| void vigra::hessianMatrixOfGaussian | ( | ... | ) |
Filter image with the 2nd derivatives of the Gaussian at the given scale to get the Hessian matrix.
The Hessian matrix is a symmetric matrix defined as:
![\[ \mbox{\rm Hessian}(I) = \left( \begin{array}{cc} G_{xx} \ast I & G_{xy} \ast I \\ G_{xy} \ast I & G_{yy} \ast I \end{array} \right) \]](form_44.png)
where 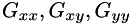 denote 2nd derivatives of Gaussians at the given scale, and
denote 2nd derivatives of Gaussians at the given scale, and 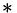 is the convolution symbol. This function calls separableConvolveX() and separableConvolveY() with the appropriate 2nd derivative of Gaussian kernels and puts the results in the three destination images. The first destination image will contain the second derivative in x-direction, the second one the mixed derivative, and the third one holds the derivative in y-direction.
is the convolution symbol. This function calls separableConvolveX() and separableConvolveY() with the appropriate 2nd derivative of Gaussian kernels and puts the results in the three destination images. The first destination image will contain the second derivative in x-direction, the second one the mixed derivative, and the third one holds the derivative in y-direction.
Function hessianOfGaussianMultiArray() performs the same filter operation on arbitrary dimensional arrays.
Declarations:
pass 2D array views:
Usage:
#include <vigra/convolution.hxx>
Namespace: vigra
| void vigra::structureTensor | ( | ... | ) |
Calculate the Structure Tensor for each pixel of and image, using Gaussian (derivative) filters.
The Structure Tensor is is a smoothed version of the Euclidean product of the gradient vector with itself. I.e. it's a symmetric matrix defined as:
![\[ \mbox{\rm StructurTensor}(I) = \left( \begin{array}{cc} G \ast (I_x I_x) & G \ast (I_x I_y) \\ G \ast (I_x I_y) & G \ast (I_y I_y) \end{array} \right) = \left( \begin{array}{cc} A & C \\ C & B \end{array} \right) \]](form_47.png)
where  denotes Gaussian smoothing at the outer scale,
denotes Gaussian smoothing at the outer scale, 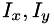 are the gradient components taken at the inner scale,
are the gradient components taken at the inner scale, 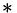 is the convolution symbol, and
is the convolution symbol, and 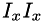 etc. are pixelwise products of the 1st derivative images. This function calls separableConvolveX() and separableConvolveY() with the appropriate Gaussian kernels and puts the results in the three separate destination images (where the first one will contain
etc. are pixelwise products of the 1st derivative images. This function calls separableConvolveX() and separableConvolveY() with the appropriate Gaussian kernels and puts the results in the three separate destination images (where the first one will contain 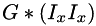 , the second one
, the second one 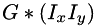 , and the third one holds
, and the third one holds 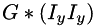 ), or into a single 3-band image (where the bands hold the result in the same order as above). The latter form is also applicable when the source image is a multi-band image (e.g. RGB). In this case, tensors are first computed for each band separately, and then summed up to get a single result tensor.
), or into a single 3-band image (where the bands hold the result in the same order as above). The latter form is also applicable when the source image is a multi-band image (e.g. RGB). In this case, tensors are first computed for each band separately, and then summed up to get a single result tensor.
Function structureTensorMultiArray() performs the same filter operation on arbitrary dimensional arrays.
Declarations:
pass 2D array views:
Usage:
#include <vigra/convolution.hxx>
Namespace: vigra
| void vigra::separableConvolveMultiArray | ( | ... | ) |
Separated convolution on multi-dimensional arrays.
This function computes a separated convolution on all dimensions of the given multi-dimensional array. Both source and destination arrays are represented by iterators, shape objects and accessors. The destination array is required to already have the correct size.
There are two variants of this functions: one takes a single kernel of type vigra::Kernel1D which is then applied to all dimensions, whereas the other requires an iterator referencing a sequence of vigra::Kernel1D objects, one for every dimension of the data. Then the first kernel in this sequence is applied to the innermost dimension (e.g. the x-axis of an image), while the last is applied to the outermost dimension (e.g. the z-axis in a 3D image).
This function may work in-place, which means that source.data() == dest.data() is allowed. A full-sized internal array is only allocated if working on the destination array directly would cause round-off errors (i.e. if typeid(typename NumericTraits<T2>::RealPromote) != typeid(T2)).
If start and stop have non-default values, they must represent a valid subarray of the input array. The convolution is then restricted to that subarray, and it is assumed that the output array only refers to the subarray (i.e. dest.shape() == stop - start). Negative ROI boundaries are interpreted relative to the end of the respective dimension (i.e. if(stop[k] < 0) stop[k] += source.shape(k);).
Declarations:
pass arbitrary-dimensional array views:
Usage:
#include <vigra/multi_convolution.hxx>
Namespace: vigra
- See Also
- vigra::Kernel1D, convolveLine()
| void vigra::convolveMultiArrayOneDimension | ( | ... | ) |
Convolution along a single dimension of a multi-dimensional arrays.
This function computes a convolution along one dimension (specified by the parameter dim of the given multi-dimensional array with the given kernel. The destination array must already have the correct size.
If start and stop have non-default values, they must represent a valid subarray of the input array. The convolution is then restricted to that subarray, and it is assumed that the output array only refers to the subarray (i.e. dest.shape() == stop - start). Negative ROI boundaries are interpreted relative to the end of the respective dimension (i.e. if(stop[k] < 0) stop[k] += source.shape(k);).
This function may work in-place, which means that source.data() == dest.data() is allowed.
Declarations:
pass arbitrary-dimensional array views:
Usage:
#include <vigra/multi_convolution.hxx>
Namespace: vigra
- See Also
- separableConvolveMultiArray()
| void vigra::gaussianSmoothMultiArray | ( | ... | ) |
Isotropic Gaussian smoothing of a multi-dimensional arrays.
This function computes an isotropic convolution of the given N-dimensional array with a Gaussian filter at the given standard deviation sigma. Both source and destination arrays are represented by iterators, shape objects and accessors. The destination array is required to already have the correct size. This function may work in-place, which means that source.data() == dest.data() is allowed. It is implemented by a call to separableConvolveMultiArray() with the appropriate kernel.
Anisotropic data should be provided with appropriate vigra::ConvolutionOptions to adjust the filter sizes for the resolution of each axis. Otherwise, the parameter opt is optional unless the parameter sigma is omitted.
If you pass vigra::BlockwiseConvolutionOptions instead, the algorithm will be executed in parallel on data blocks of a certain size. The block size can be customized via BlockwiseConvolutionOptions::blockShape(), but the defaults usually work reasonably. By default, the number of threads equals the capabilities of your hardware, but you can change this via BlockwiseConvolutionOptions::numThreads().
Declarations:
pass arbitrary-dimensional array views:
Usage:
#include <vigra/multi_convolution.hxx> (sequential version)
#include <vigra/multi_blockwise.hxx> (parallel version)
Namespace: vigra
Multi-threaded execution:
Usage with anisotropic data:
- See Also
- separableConvolveMultiArray()
| void vigra::gaussianGradientMultiArray | ( | ... | ) |
Calculate Gaussian gradient of a multi-dimensional arrays.
This function computes the Gaussian gradient of the given N-dimensional array with a sequence of first-derivative-of-Gaussian filters at the given standard deviation sigma (differentiation is applied to each dimension in turn, starting with the innermost dimension). The destination array is required to have a vector valued pixel type with as many elements as the number of dimensions. This function is implemented by calls to separableConvolveMultiArray() with the appropriate kernels.
Anisotropic data should be provided with appropriate vigra::ConvolutionOptions to adjust the filter sizes for the resolution of each axis. Otherwise, the parameter opt is optional unless the parameter sigma is omitted.
If you pass vigra::BlockwiseConvolutionOptions instead, the algorithm will be executed in parallel on data blocks of a certain size. The block size can be customized via BlockwiseConvolutionOptions::blockShape(), but the defaults usually work reasonably. By default, the number of threads equals the capabilities of your hardware, but you can change this via BlockwiseConvolutionOptions::numThreads().
Declarations:
pass arbitrary-dimensional array views:
Usage:
#include <vigra/multi_convolution.hxx> (sequential version)
#include <vigra/multi_blockwise.hxx> (parallel version)
Namespace: vigra
Usage with anisotropic data:
- See Also
- separableConvolveMultiArray()
| void vigra::symmetricGradientMultiArray | ( | ... | ) |
Calculate gradient of a multi-dimensional arrays using symmetric difference filters.
This function computes the gradient of the given N-dimensional array with a sequence of symmetric difference filters a (differentiation is applied to each dimension in turn, starting with the innermost dimension). The destination array is required to have a vector valued pixel type with as many elements as the number of dimensions. This function is implemented by calls to convolveMultiArrayOneDimension() with the symmetric difference kernel.
Anisotropic data should be provided with appropriate vigra::ConvolutionOptions to adjust the filter sizes for the resolution of each axis. Otherwise, the parameter opt is optional unless the parameter sigma is omitted.
If you pass vigra::BlockwiseConvolutionOptions instead, the algorithm will be executed in parallel on data blocks of a certain size. The block size can be customized via BlockwiseConvolutionOptions::blockShape(), but the defaults usually work reasonably. By default, the number of threads equals the capabilities of your hardware, but you can change this via BlockwiseConvolutionOptions::numThreads().
Declarations:
pass arbitrary-dimensional array views:
Usage:
#include <vigra/multi_convolution.hxx> (sequential version)
#include <vigra/multi_blockwise.hxx> (parallel version)
Namespace: vigra
Usage with anisotropic data:
- See Also
- convolveMultiArrayOneDimension()
| void vigra::laplacianOfGaussianMultiArray | ( | ... | ) |
Calculate Laplacian of a N-dimensional arrays using Gaussian derivative filters.
This function computes the Laplacian of the given N-dimensional array with a sequence of second-derivative-of-Gaussian filters at the given standard deviation sigma. Both source and destination arrays must have scalar value_type. This function is implemented by calls to separableConvolveMultiArray() with the appropriate kernels, followed by summation.
Anisotropic data should be provided with appropriate vigra::ConvolutionOptions to adjust the filter sizes for the resolution of each axis. Otherwise, the parameter opt is optional unless the parameter sigma is omitted.
If you pass vigra::BlockwiseConvolutionOptions instead, the algorithm will be executed in parallel on data blocks of a certain size. The block size can be customized via BlockwiseConvolutionOptions::blockShape(), but the defaults usually work reasonably. By default, the number of threads equals the capabilities of your hardware, but you can change this via BlockwiseConvolutionOptions::numThreads().
Declarations:
pass arbitrary-dimensional array views:
Usage:
#include <vigra/multi_convolution.hxx> (sequential version)
#include <vigra/multi_blockwise.hxx> (parallel version)
Namespace: vigra
Usage with anisotropic data:
- See Also
- separableConvolveMultiArray()
| void vigra::gaussianDivergenceMultiArray | ( | ... | ) |
Calculate the divergence of a vector field using Gaussian derivative filters.
This function computes the divergence of the given N-dimensional vector field with a sequence of first-derivative-of-Gaussian filters at the given standard deviation sigma. The input vector field can either be given as a sequence of scalar array views (one for each vector field component), represented by an iterator range, or by a single vector array with the appropriate shape. This function is implemented by calls to separableConvolveMultiArray() with the suitable kernels, followed by summation.
Anisotropic data should be provided with appropriate vigra::ConvolutionOptions to adjust the filter sizes for the resolution of each axis. Otherwise, the parameter opt is optional unless the parameter sigma is omitted.
If you pass vigra::BlockwiseConvolutionOptions instead, the algorithm will be executed in parallel on data blocks of a certain size. The block size can be customized via BlockwiseConvolutionOptions::blockShape(), but the defaults usually work reasonably. By default, the number of threads equals the capabilities of your hardware, but you can change this via BlockwiseConvolutionOptions::numThreads().
Declarations:
pass arbitrary-dimensional array views:
Usage:
#include <vigra/multi_convolution.hxx> (sequential version)
#include <vigra/multi_blockwise.hxx> (parallel version)
Namespace: vigra
Usage with anisotropic data:
| void vigra::hessianOfGaussianMultiArray | ( | ... | ) |
Calculate Hessian matrix of a N-dimensional arrays using Gaussian derivative filters.
This function computes the Hessian matrix the given scalar N-dimensional array with a sequence of second-derivative-of-Gaussian filters at the given standard deviation sigma. The destination array must have a vector valued element type with N*(N+1)/2 elements (it represents the upper triangular part of the symmetric Hessian matrix, flattened row-wise). This function is implemented by calls to separableConvolveMultiArray() with the appropriate kernels.
Anisotropic data should be provided with appropriate vigra::ConvolutionOptions to adjust the filter sizes for the resolution of each axis. Otherwise, the parameter opt is optional unless the parameter sigma is omitted.
If you pass vigra::BlockwiseConvolutionOptions instead, the algorithm will be executed in parallel on data blocks of a certain size. The block size can be customized via BlockwiseConvolutionOptions::blockShape(), but the defaults usually work reasonably. By default, the number of threads equals the capabilities of your hardware, but you can change this via BlockwiseConvolutionOptions::numThreads().
Declarations:
pass arbitrary-dimensional array views:
Usage:
#include <vigra/multi_convolution.hxx> (sequential version)
#include <vigra/multi_blockwise.hxx> (parallel version)
Namespace: vigra
Usage with anisotropic data:
| void vigra::structureTensorMultiArray | ( | ... | ) |
Calculate the structure tensor of a multi-dimensional arrays.
This function computes the gradient (outer product) tensor for each element of the given N-dimensional array with first-derivative-of-Gaussian filters at the given innerScale, followed by Gaussian smoothing at outerScale. The destination array must have a vector valued pixel type with N*(N+1)/2 elements (it represents the upper triangular part of the symmetric structure tensor matrix, flattened row-wise). If the source array is also vector valued, the resulting structure tensor is the sum of the individual tensors for each channel. This function is implemented by calls to separableConvolveMultiArray() with the appropriate kernels.
Anisotropic data should be provided with appropriate vigra::ConvolutionOptions to adjust the filter sizes for the resolution of each axis. Otherwise, the parameter opt is optional unless the parameters innerScale and outerScale are both omitted.
If you pass vigra::BlockwiseConvolutionOptions instead, the algorithm will be executed in parallel on data blocks of a certain size. The block size can be customized via BlockwiseConvolutionOptions::blockShape(), but the defaults usually work reasonably. By default, the number of threads equals the capabilities of your hardware, but you can change this via BlockwiseConvolutionOptions::numThreads().
Declarations:
pass arbitrary-dimensional array views:
Usage:
#include <vigra/multi_convolution.hxx> (sequential version)
#include <vigra/multi_blockwise.hxx> (parallel version)
Namespace: vigra
Usage with anisotropic data:
| void vigra::convolveFFT | ( | ... | ) |
Convolve an array with a kernel by means of the Fourier transform.
Thanks to the convolution theorem of Fourier theory, a convolution in the spatial domain is equivalent to a multiplication in the frequency domain. Thus, for certain kernels (especially large, non-separable ones), it is advantageous to perform the convolution by first transforming both array and kernel to the frequency domain, multiplying the frequency representations, and transforming the result back into the spatial domain. Some kernels have a much simpler definition in the frequency domain, so that they are readily computed there directly, avoiding Fourier transformation of those kernels.
The following functions implement various variants of FFT-based convolution:
- convolveFFT
- Convolve a real-valued input array with a kernel such that the result is also real-valued. That is, the kernel is either provided as a real-valued array in the spatial domain, or as a complex-valued array in the Fourier domain, using the half-space format of the R2C Fourier transform (see below).
- convolveFFTMany
- Like
convolveFFT, but you may provide many kernels at once (using an iterator pair specifying the kernel sequence). This has the advantage that the forward transform of the input array needs to be executed only once. - convolveFFTComplex
- Convolve a complex-valued input array with a complex-valued kernel, resulting in a complex-valued output array. An additional flag is used to specify whether the kernel is defined in the spatial or frequency domain.
- convolveFFTComplexMany
- Like
convolveFFTComplex, but you may provide many kernels at once (using an iterator pair specifying the kernel sequence). This has the advantage that the forward transform of the input array needs to be executed only once.
The output arrays must have the same shape as the input arrays. In the "Many" variants of the convolution functions, the kernels must all have the same shape.
The origin of the kernel is always assumed to be in the center of the kernel array (precisely, at the point floor(kernel.shape() / 2.0), except when the half-space format is used, see below). The function moveDCToUpperLeft() will be called internally to align the kernel with the transformed input as appropriate.
If a real input is combined with a real kernel, the kernel is automatically assumed to be defined in the spatial domain. If a real input is combined with a complex kernel, the kernel is assumed to be defined in the Fourier domain in half-space format. If the input array is complex, a flag fourierDomainKernel determines where the kernel is defined.
When the kernel is defined in the spatial domain, the convolution functions will automatically pad (enlarge) the input array by at least the kernel radius in each direction. The newly added space is filled according to reflective boundary conditions in order to minimize border artifacts during convolution. It is thus ensured that convolution in the Fourier domain yields the same results as convolution in the spatial domain (e.g. when separableConvolveMultiArray() is called with the same kernel). A little further padding may be added to make sure that the padded array shape uses integers which have only small prime factors, because FFTW is then able to use the fastest possible algorithms. Any padding is automatically removed from the result arrays before the function returns.
When the kernel is defined in the frequency domain, it must be complex-valued, and its shape determines the shape of the Fourier representation (i.e. the input is padded according to the shape of the kernel). If we are going to perform a complex-valued convolution, the kernel must be defined for the entire frequency domain, and its shape directly determines the size of the FFT.
In contrast, a frequency domain kernel for a real-valued convolution must have symmetry properties that allow to drop half of the kernel coefficients, as in the R2C transform. That is, the kernel must have the half-space format, that is the shape returned by fftwCorrespondingShapeR2C(fourier_shape), where fourier_shape is the desired logical shape of the frequency representation (and thus the size of the padded input). The origin of the kernel must be at the point (0, floor(fourier_shape[0] / 2.0), ..., floor(fourier_shape[N-1] / 2.0)) (i.e. as in a regular kernel except for the first dimension).
The Real type in the declarations can be double, float, and long double. Your program must always link against libfftw3. If you use float or long double arrays, you must additionally link against libfftw3f and libfftw3l respectively.
The Fourier transform functions internally create FFTW plans which control the algorithm details. The plans are created with the flag FFTW_ESTIMATE, i.e. optimal settings are guessed or read from saved "wisdom" files. If you need more control over planning, you can use the class FFTWConvolvePlan.
See also applyFourierFilter() for corresponding functionality on the basis of the old image iterator interface.
Declarations:
Real-valued convolution with kernel in the spatial domain:
Real-valued convolution with kernel in the Fourier domain (half-space format):
Series of real-valued convolutions with kernels in the spatial or Fourier domain (the kernel and out sequences must have the same length):
Complex-valued convolution (parameter fourierDomainKernel determines if the kernel is defined in the spatial or Fourier domain):
Series of complex-valued convolutions (parameter fourierDomainKernel determines if the kernels are defined in the spatial or Fourier domain, the kernel and out sequences must have the same length):
Usage:
#include <vigra/multi_fft.hxx>
Namespace: vigra
| void vigra::convolveFFTComplex | ( | ... | ) |
Convolve a complex-valued array by means of the Fourier transform.
See convolveFFT() for details.
| void vigra::convolveFFTMany | ( | ... | ) |
Convolve a real-valued array with a sequence of kernels by means of the Fourier transform.
See convolveFFT() for details.
| void vigra::convolveFFTComplexMany | ( | ... | ) |
Convolve a complex-valued array with a sequence of kernels by means of the Fourier transform.
See convolveFFT() for details.
| void vigra::correlateFFT | ( | ... | ) |
Correlate an array with a kernel by means of the Fourier transform.
This function correlates a real-valued input array with a real-valued kernel such that the result is also real-valued. Thanks to the correlation theorem of Fourier theory, a correlation in the spatial domain is equivalent to a multiplication with the complex conjugate in the frequency domain. Thus, for certain kernels (especially large, non-separable ones), it is advantageous to perform the correlation by first transforming both array and kernel to the frequency domain, multiplying the frequency representations, and transforming the result back into the spatial domain.
The output arrays must have the same shape as the input arrays.
See also convolveFFT() for corresponding functionality.
Declarations:
Usage:
#include <vigra/multi_fft.hxx>
Namespace: vigra
| void vigra::normalizedConvolveImage | ( | ... | ) |
Performs a 2-dimensional normalized convolution, i.e. convolution with a mask image.
This functions computes normalized convolution as defined in Knutsson, H. and Westin, C-F.: Normalized and differential convolution: Methods for Interpolation and Filtering of incomplete and uncertain data. Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, 1993, 515-523.
The mask image must be binary and encodes which pixels of the original image are valid. It is used as follows: Only pixel under the mask are used in the calculations. Whenever a part of the kernel lies outside the mask, it is ignored, and the kernel is renormalized to its original norm (analogous to the CLIP BorderTreatmentMode). Thus, a useful convolution result is computed whenever at least one valid pixel is within the current window Thus, destination pixels not under the mask still receive a value if they are near the mask. Therefore, this algorithm is useful as an interpolator of sparse input data. If you are only interested in the destination values under the mask, you can perform a subsequent copyImageIf().
The KernelIterator must point to the center of the kernel, and the kernel's size is given by its upper left (x and y of distance <= 0) and lower right (distance >= 0) corners. The image must always be larger than the kernel. At those positions where the kernel does not completely fit into the image, the specified BorderTreatmentMode is applied. Only BORDER_TREATMENT_CLIP and BORDER_TREATMENT_AVOID are currently supported.
The images's pixel type (SrcAccessor::value_type) must be a linear space over the kernel's value_type (KernelAccessor::value_type), i.e. addition of source values, multiplication with kernel values, and NumericTraits must be defined. The kernel's value_type must be an algebraic field, i.e. the arithmetic operations (+, -, *, /) and NumericTraits must be defined.
Declarations:
pass 2D array views:
Usage:
#include <vigra/stdconvolution.hxx>
Namespace: vigra
Preconditions:
-
The image must be longer than the kernel radius:
w > std::max(kernel.lowerRight().x, -kernel.upperLeft().x)andh > std::max(kernel.lowerRight().y, -kernel.upperLeft().y). - The sum of kernel elements must be != 0.
-
border == BORDER_TREATMENT_CLIP || border == BORDER_TREATMENT_AVOID
| void vigra::convolveImageWithMask | ( | ... | ) |
Deprecated name of 2-dimensional normalized convolution, i.e. convolution with a mask image.
See normalizedConvolveImage() for documentation.
Declarations:
pass 2D array views: