
|
Optimization and Regression |  |
Classes | |
| class | LeastAngleRegressionOptions |
| Pass options to leastAngleRegression(). More... | |
| class | NonlinearLSQOptions |
| Pass options to nonlinearLeastSquares(). More... | |
Functions | |
| template<... > | |
| unsigned int | leastAngleRegression (...) |
| template<class T , class C1 , class C2 , class C3 > | |
| bool | leastSquares (MultiArrayView< 2, T, C1 > const &A, MultiArrayView< 2, T, C2 > const &b, MultiArrayView< 2, T, C3 > &x, std::string method="QR") |
| template<... > | |
| void | nonlinearLeastSquares (...) |
| Fit a non-linear model to given data by minimizing least squares loss. More... | |
| template<... > | |
| unsigned int | nonnegativeLeastSquares (...) |
| template<... > | |
| unsigned int | quadraticProgramming (...) |
| template<class T , class C1 , class C2 , class C3 > | |
| bool | ridgeRegression (MultiArrayView< 2, T, C1 > const &A, MultiArrayView< 2, T, C2 > const &b, MultiArrayView< 2, T, C3 > &x, double lambda) |
| template<class T , class C1 , class C2 , class C3 , class Array > | |
| bool | ridgeRegressionSeries (MultiArrayView< 2, T, C1 > const &A, MultiArrayView< 2, T, C2 > const &b, MultiArrayView< 2, T, C3 > &x, Array const &lambda) |
| template<class T , class C1 , class C2 , class C3 , class C4 > | |
| bool | weightedLeastSquares (MultiArrayView< 2, T, C1 > const &A, MultiArrayView< 2, T, C2 > const &b, MultiArrayView< 2, T, C3 > const &weights, MultiArrayView< 2, T, C4 > &x, std::string method="QR") |
| template<class T , class C1 , class C2 , class C3 , class C4 > | |
| bool | weightedRidgeRegression (MultiArrayView< 2, T, C1 > const &A, MultiArrayView< 2, T, C2 > const &b, MultiArrayView< 2, T, C3 > const &weights, MultiArrayView< 2, T, C4 > &x, double lambda) |
Detailed Description
Function Documentation
| unsigned int vigra::quadraticProgramming | ( | ... | ) |
Solve Quadratic Programming Problem.
The quadraticProgramming() function implements the algorithm described in
D. Goldfarb, A. Idnani: "A numerically stable dual method for solving strictly convex quadratic programs", Mathematical Programming 27:1-33, 1983.
for the solution of (convex) quadratic programming problems by means of a primal-dual method.
#include <vigra/quadprog.hxx>
Namespaces: vigra
Declaration:
The problem must be specified in the form:
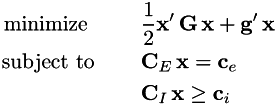
Matrix G G must be symmetric positive definite, and matrix CE must have full row rank. Matrix and vector dimensions must be as follows:
- G: [n * n], g: [n * 1]
- CE: [me * n], ce: [me * 1]
- CI: [mi * n], ci: [mi * 1]
- x: [n * 1]
The function writes the optimal solution into the vector x and returns the cost of this solution. If the problem is infeasible, std::numeric_limits::infinity() is returned. In this case the value of vector x is undefined.
Usage:
Minimize f = 0.5 * x'*G*x + g'*x subject to -1 <= x <= 1. The solution is x' = [1.0, 0.5, -1.0] with f = -22.625.
This algorithm can also be used to solve non-negative least squares problems (see nonnegativeLeastSquares() for an alternative algorithm). Consider the problem to minimize f = (A*x - b)' * (A*x - b) subject to x >= 0. Expanding the product in the objective gives f = x'*A'*A*x - 2*b'*A*x + b'*b. This is equivalent to the problem of minimizing fn = 0.5 * x'*G*x + g'*x with G = A'*A and g = -A'*b (the constant term b'*b has no effect on the optimal solution and can be dropped). The following code computes the solution x' = [18.4493, 0, 4.50725]:
- Examples:
- nnlsq.cxx.
| bool vigra::linalg::leastSquares | ( | MultiArrayView< 2, T, C1 > const & | A, |
| MultiArrayView< 2, T, C2 > const & | b, | ||
| MultiArrayView< 2, T, C3 > & | x, | ||
| std::string | method = "QR" |
||
| ) |
Ordinary Least Squares Regression.
Given a matrix A with m rows and n columns (with m >= n), and a column vector b of length m rows, this function computes the column vector x of length n rows that solves the optimization problem
![\[ \tilde \textrm{\bf x} = \textrm{argmin} \left|\left|\textrm{\bf A} \textrm{\bf x} - \textrm{\bf b}\right|\right|_2^2 \]](form_75.png)
When b is a matrix with k columns, x must also have k columns, which will contain the solutions for the corresponding columns of b. Note that all matrices must already have the correct shape.
This function is just another name for linearSolve(), perhaps leading to more readable code when A is a rectangular matrix. It returns false when the rank of A is less than n. See linearSolve() for more documentation.
#include <vigra/regression.hxx>
Namespaces: vigra and vigra::linalg
| bool vigra::linalg::weightedLeastSquares | ( | MultiArrayView< 2, T, C1 > const & | A, |
| MultiArrayView< 2, T, C2 > const & | b, | ||
| MultiArrayView< 2, T, C3 > const & | weights, | ||
| MultiArrayView< 2, T, C4 > & | x, | ||
| std::string | method = "QR" |
||
| ) |
Weighted Least Squares Regression.
Given a matrix A with m rows and n columns (with m >= n), a vector b of length m, and a weight vector weights of length m with non-negative entries, this function computes the vector x of length n that solves the optimization problem
![\[ \tilde \textrm{\bf x} = \textrm{argmin} \left(\textrm{\bf A} \textrm{\bf x} - \textrm{\bf b}\right)^T \textrm{diag}(\textrm{\bf weights}) \left(\textrm{\bf A} \textrm{\bf x} - \textrm{\bf b}\right) \]](form_76.png)
where diag(weights) creates a diagonal matrix from weights. The algorithm calls leastSquares() on the equivalent problem
![\[ \tilde \textrm{\bf x} = \textrm{argmin} \left|\left|\textrm{diag}(\textrm{\bf weights})^{1/2}\textrm{\bf A} \textrm{\bf x} - \textrm{diag}(\textrm{\bf weights})^{1/2} \textrm{\bf b}\right|\right|_2^2 \]](form_77.png)
where the square root of weights is just taken element-wise.
When b is a matrix with k columns, x must also have k columns, which will contain the solutions for the corresponding columns of b. Note that all matrices must already have the correct shape.
The function returns false when the rank of the weighted matrix A is less than n.
#include <vigra/regression.hxx>
Namespaces: vigra and vigra::linalg
| bool vigra::linalg::ridgeRegression | ( | MultiArrayView< 2, T, C1 > const & | A, |
| MultiArrayView< 2, T, C2 > const & | b, | ||
| MultiArrayView< 2, T, C3 > & | x, | ||
| double | lambda | ||
| ) |
Ridge Regression.
Given a matrix A with m rows and n columns (with m >= n), a vector b of length m, and a regularization parameter lambda >= 0.0, this function computes the vector x of length n that solves the optimization problem
![\[ \tilde \textrm{\bf x} = \textrm{argmin} \left|\left|\textrm{\bf A} \textrm{\bf x} - \textrm{\bf b}\right|\right|_2^2 + \lambda \textrm{\bf x}^T\textrm{\bf x} \]](form_78.png)
This is implemented by means of singularValueDecomposition().
When b is a matrix with k columns, x must also have k columns, which will contain the solutions for the corresponding columns of b. Note that all matrices must already have the correct shape.
The function returns false if the rank of A is less than n and lambda == 0.0.
#include <vigra/regression.hxx>
Namespaces: vigra and vigra::linalg
| bool vigra::linalg::weightedRidgeRegression | ( | MultiArrayView< 2, T, C1 > const & | A, |
| MultiArrayView< 2, T, C2 > const & | b, | ||
| MultiArrayView< 2, T, C3 > const & | weights, | ||
| MultiArrayView< 2, T, C4 > & | x, | ||
| double | lambda | ||
| ) |
Weighted ridge Regression.
Given a matrix A with m rows and n columns (with m >= n), a vector b of length m, a weight vector weights of length m with non-negative entries, and a regularization parameter lambda >= 0.0 this function computes the vector x of length n that solves the optimization problem
![\[ \tilde \textrm{\bf x} = \textrm{argmin} \left(\textrm{\bf A} \textrm{\bf x} - \textrm{\bf b}\right)^T \textrm{diag}(\textrm{\bf weights}) \left(\textrm{\bf A} \textrm{\bf x} - \textrm{\bf b}\right) + \lambda \textrm{\bf x}^T\textrm{\bf x} \]](form_79.png)
where diag(weights) creates a diagonal matrix from weights. The algorithm calls ridgeRegression() on the equivalent problem
![\[ \tilde \textrm{\bf x} = \textrm{argmin} \left|\left|\textrm{diag}(\textrm{\bf weights})^{1/2}\textrm{\bf A} \textrm{\bf x} - \textrm{diag}(\textrm{\bf weights})^{1/2} \textrm{\bf b}\right|\right|_2^2 + \lambda \textrm{\bf x}^T\textrm{\bf x} \]](form_80.png)
where the square root of weights is just taken element-wise. This solution is computed by means of singularValueDecomposition().
When b is a matrix with k columns, x must also have k columns, which will contain the solutions for the corresponding columns of b. Note that all matrices must already have the correct shape.
The function returns false if the rank of A is less than n and lambda == 0.0.
#include <vigra/regression.hxx>
Namespaces: vigra and vigra::linalg
| bool vigra::linalg::ridgeRegressionSeries | ( | MultiArrayView< 2, T, C1 > const & | A, |
| MultiArrayView< 2, T, C2 > const & | b, | ||
| MultiArrayView< 2, T, C3 > & | x, | ||
| Array const & | lambda | ||
| ) |
Ridge Regression with many lambdas.
This executes ridgeRegression() for a sequence of regularization parameters. This is implemented so that the singularValueDecomposition() has to be executed only once. lambda must be an array conforming to the std::vector interface, i.e. must support lambda.size() and lambda[k]. The columns of the matrix x will contain the solutions for the corresponding lambda, so the number of columns of the matrix x must be equal to lambda.size(), and b must be a columns vector, i.e. cannot contain several right hand sides at once.
The function returns false when the matrix A is rank deficient. If this happens, and one of the lambdas is zero, the corresponding column of x will be skipped.
#include <vigra/regression.hxx>
Namespaces: vigra and vigra::linalg
| unsigned int vigra::linalg::leastAngleRegression | ( | ... | ) |
Least Angle Regression.
#include <vigra/regression.hxx>
Namespaces: vigra and vigra::linalg
Declarations:
This function implements Least Angle Regression (LARS) as described in
B.Efron, T.Hastie, I.Johnstone, and R.Tibshirani: "Least Angle Regression", Annals of Statistics 32(2):407-499, 2004.
It is an efficient algorithm to solve the L1-regularized least squares (LASSO) problem
![\[ \tilde \textrm{\bf x} = \textrm{argmin} \left|\left|\textrm{\bf A} \textrm{\bf x} - \textrm{\bf b}\right|\right|_2^2 \textrm{ subject to } \left|\left|\textrm{\bf x}\right|\right|_1\le s \]](form_81.png)
and the L1-regularized non-negative least squares (NN-LASSO) problem
![\[ \tilde \textrm{\bf x} = \textrm{argmin} \left|\left|\textrm{\bf A} \textrm{\bf x} - \textrm{\bf b}\right|\right|_2^2 \textrm{ subject to } \left|\left|\textrm{\bf x}\right|\right|_1\le s \textrm{ and } \textrm{\bf x}\ge \textrm{\bf 0} \]](form_82.png)
where A is a matrix with m rows and n columns (often with m < n), b a vector of length m, and a regularization parameter s >= 0.0. L1-regularization has the desirable effect that it causes the solution x to be sparse, i.e. only the most important elements in x (called the active set) have non-zero values. The key insight of the LARS algorithm is the following: When the solution vector x is considered as a function of the regularization parameter s, then x(s) is a piecewise linear function, i.e. a polyline in n-dimensional space. The knots of the polyline x(s) are located precisely at those values of s where one variable enters or leaves the active set and can be efficiently computed.
Therefore, leastAngleRegression() returns the entire solution path as a sequence of knot points, starting at 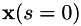 (where the only feasible solution is obviously x = 0) and ending at
(where the only feasible solution is obviously x = 0) and ending at 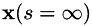 (where the solution becomes the ordinary least squares solution). Actually, the initial null solution is not explicitly returned, i.e. the sequence starts at the first non-zero solution with one variable in the active set. The function leastAngleRegression() returns the number of solutions (i.e. knot points) computed.
(where the solution becomes the ordinary least squares solution). Actually, the initial null solution is not explicitly returned, i.e. the sequence starts at the first non-zero solution with one variable in the active set. The function leastAngleRegression() returns the number of solutions (i.e. knot points) computed.
The sequences of active sets and corresponding variable weights are returned in activeSets and solutions respectively. That is, activeSets[i] is an ArrayVector<int> containing the indices of the variables that are active at the i-th knot, and solutions is a Matrix<T> containing the weights of those variables, in the same order (see example below). Variables not contained in activeSets[i] are zero at this solution.
The behavior of the algorithm can be adapted by LeastAngleRegressionOptions:
- options.lasso() (active by default)
- Compute the LASSO solution as described above.
- options.nnlasso() (inactive by default)
- Compute non-negative LASSO solutions, i.e. use the additional constraint that x >= 0 in all solutions.
- options.lars() (inactive by default)
- Compute a solution path according to the plain LARS rule, i.e. never remove a variable from the active set once it entered.
- options.leastSquaresSolutions(bool) (default: true)
- Use the algorithm mode selected above to determine the sequence of active sets, but then compute and return an ordinary (unconstrained) least squares solution for every active set.
Note: The second form of leastAngleRegression() ignores this option and does always compute both constrained and unconstrained solutions (returned in lasso_solutions and lsq_solutions respectively). - maxSolutionCount(unsigned int n) (default: n = 0, i.e. compute all solutions)
- Compute at most
nsolutions.
Usage:
Required Interface:
-
Tmust be numeric type (compatible to double) -
Array1 a1;
a1.push_back(ArrayVector<int>()); -
Array2 a2;
a2.push_back(Matrix<T>());
| unsigned int vigra::linalg::nonnegativeLeastSquares | ( | ... | ) |
Non-negative Least Squares Regression.
Given a matrix A with m rows and n columns (with m >= n), and a column vector b of length m rows, this function computes a column vector x of length n with non-negative entries that solves the optimization problem
![\[ \tilde \textrm{\bf x} = \textrm{argmin} \left|\left|\textrm{\bf A} \textrm{\bf x} - \textrm{\bf b}\right|\right|_2^2 \textrm{ subject to } \textrm{\bf x} \ge \textrm{\bf 0} \]](form_85.png)
Both b and x must be column vectors (i.e. matrices with 1 column). Note that all matrices must already have the correct shape. The solution is computed by means of leastAngleRegression() with non-negativity constraint.
#include <vigra/regression.hxx>
Namespaces: vigra and vigra::linalg
Declarations:
- Examples:
- nnlsq.cxx.
| void vigra::nonlinearLeastSquares | ( | ... | ) |
Fit a non-linear model to given data by minimizing least squares loss.
Declarations:
This function implements the Levenberg-Marquardt algorithm to fit a non-linear model to given data. The model depends on a vector of parameters p which are to be choosen such that the least-squares residual between the data and the model's predictions is minimized according to the objective function:
![\[ \tilde \textrm{\bf p} = \textrm{argmin}_{\textrm{\bf p}} \sum_i \left( y_i - f(\textrm{\bf x}_i; \textrm{\bf p}) \right)^2 \]](form_133.png)
where  is the model to be optimized (with arguments
is the model to be optimized (with arguments 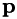 and parameters
and parameters 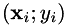 ), and
), and 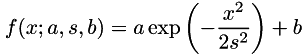 are the feature/response pairs of the given data. Since the model is non-linear (otherwise, you should use ordinary leastSquares()), it must be linearized in terms of a first-order Taylor expansion, and the optimal parameters p have to be determined iteratively. In order for the iterations to converge to the desired solution, a good initial guess on p is required.
are the feature/response pairs of the given data. Since the model is non-linear (otherwise, you should use ordinary leastSquares()), it must be linearized in terms of a first-order Taylor expansion, and the optimal parameters p have to be determined iteratively. In order for the iterations to converge to the desired solution, a good initial guess on p is required.
The model must be specified by a functor which takes one of the following forms:
Each call to the functor's operator() computes the model's prediction for a single data point. The current model parameters are specified in a TinyVector of appropriate length. The type T must be templated: normally, it is the same as DataType, but the nonlinearLeastSquares() function will temporarily replace it with a special number type that supports automatic differentiation (see vigra::autodiff::DualVector). In this way, the derivatives needed in the model's Taylor expansion can be computed automatically.
When the model is univariate (has a single scalar argument), the samples must be passed to nonlinearLeastSquares() in a pair 'x', 'y' of 1D MultiArrayViews (variant 1). When the model is multivariate (has a vector-valued argument), the 'x' input must be a 2D MultiArrayView (variant 2) whose rows represent a single data sample (i.e. the number of columns corresponds to the length of the model's argument vector). The number of rows in 'x' defines the number of data samples and must match the length of array 'y'.
The TinyVector 'model_parameters' holds the initial guess for the model parameters and will be overwritten by the optimal values found by the algorithm. The algorithm's internal behavior can be controlled by customizing the option object vigra::NonlinearLSQOptions.
The function returns the residual sum of squared errors of the final solution.
Usage:
#include <vigra/regression.hxx>
Namespace: vigra
Suppose that we want to fit a centered Gaussian function of the form
![\[ f(x ; a, s, b) = a \exp\left(-\frac{x^2}{2 s^2}\right) + b \]](form_138.png)
to noisy data 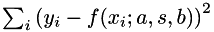 , i.e. we want to find parameters a, s, b such that the residual
, i.e. we want to find parameters a, s, b such that the residual 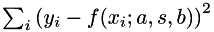 is minimized. The model parameters are placed in a
is minimized. The model parameters are placed in a TinyVector<T, 3> p according to the rules
p[0] <=> a, p[1] <=> s and p[2] <=> b.
The following functor computes the model's prediction for a single data point x:
Now we can find optimal values for the parameters like this: