
|
Non-linear Diffusion and Total Variation |  |
Classes | |
| class | DiffusivityFunctor< Value > |
| Diffusivity functor for non-linear diffusion. More... | |
Functions | |
| template<... > | |
| void | anisotropicTotalVariationFilter (...) |
| Performs Anisotropic Total Variation Regularization. More... | |
| template<... > | |
| void | getAnisotropy (...) |
| Sets up directional data for anisotropic regularization. More... | |
| template<... > | |
| void | nonlinearDiffusion (...) |
| Perform edge-preserving smoothing at the given scale. More... | |
| template<... > | |
| void | nonlinearDiffusionExplicit (...) |
| Perform edge-preserving smoothing at the given scale using an explicit scheme. More... | |
| template<... > | |
| void | secondOrderTotalVariationFilter (...) |
| Performs Anisotropic Total Variation Regularization. More... | |
| template<... > | |
| void | totalVariationFilter (...) |
| Performs standard Total Variation Regularization. More... | |
Detailed Description
Perform edge-preserving smoothing.
Function Documentation
| void vigra::nonlinearDiffusion | ( | ... | ) |
Perform edge-preserving smoothing at the given scale.
The algorithm solves the non-linear diffusion equation
![\[ \frac{\partial}{\partial t} u = \frac{\partial}{\partial x} \left( g(|\nabla u|) \frac{\partial}{\partial x} u \right) \]](form_70.png)
where t is the time, x is the location vector, u( x , t) is the smoothed image at time t, and g(.) is the location dependent diffusivity. At time zero, the image u( x , 0) is simply the original image. The time is proportional to the square of the scale parameter: 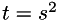 . The diffusion equation is solved iteratively according to the Additive Operator Splitting Scheme (AOS) from
. The diffusion equation is solved iteratively according to the Additive Operator Splitting Scheme (AOS) from
J. Weickert: "Recursive Separable Schemes for Nonlinear Diffusion Filters", in: B. ter Haar Romeny, L. Florack, J. Koenderingk, M. Viergever (eds.): 1st Intl. Conf. on Scale-Space Theory in Computer Vision 1997, Springer LNCS 1252
DiffusivityFunctor implements the gradient-dependent local diffusivity. It is passed as an argument to gradientBasedTransform(). The return value must be between 0 and 1 and determines the weight a pixel gets when its neighbors are smoothed. Weickert recommends the use of the diffusivity implemented by class DiffusivityFunctor. It's also possible to use other functors, for example one that always returns 1, in which case we obtain the solution to the linear diffusion equation, i.e. Gaussian convolution.
The source value type must be a linear space with internal addition, scalar multiplication, and NumericTraits defined. The value_type of the DiffusivityFunctor must be the scalar field over wich the source value type's linear space is defined.
In addition to nonlinearDiffusion(), there is an algorithm nonlinearDiffusionExplicit() which implements the Explicit Scheme described in the above article. Both algorithms have the same interface, but the explicit scheme gives slightly more accurate approximations of the diffusion process at the cost of much slower processing.
Declarations:
pass 2D array views:
Usage:
#include <vigra/nonlineardiffusion.hxx>
Namespace: vigra
Precondition:
scale > 0
- See Also
- vigra::DiffusivityFunctor
- Examples:
- smooth.cxx.
| void vigra::nonlinearDiffusionExplicit | ( | ... | ) |
Perform edge-preserving smoothing at the given scale using an explicit scheme.
See nonlinearDiffusion().
| void vigra::totalVariationFilter | ( | ... | ) |
Performs standard Total Variation Regularization.
The algorithm minimizes
![\[ \min_u \int_\Omega \frac{1}{2} (u-f)^2\;dx + \alpha TV(u)\qquad\qquad (1) \]](form_93.png)
where 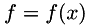 are the two dimensional noisy data,
are the two dimensional noisy data, 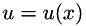 are the smoothed data,
are the smoothed data, 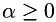 is the filter parameter and
is the filter parameter and 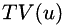 is the total variation semi-norm.
is the total variation semi-norm.
Declarations:
totalVariationFilter() implements a primal-dual algorithm to solve (1).
Input:
| data: | input data to be smoothed. |
| alpha: | smoothing parameter. |
| steps: | maximal number of iteration steps. |
| eps: | The algorithm stops, if the primal-dual gap is below the threshold eps. |
Output:
out contains the filtered data.
In addition, a point-wise weight ( 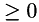 ) for the data term can be provided (overloaded function).
) for the data term can be provided (overloaded function).
Usage:
#include <vigra/tv_filter.hxx>
or
- Examples:
- total_variation.cxx.
| void vigra::getAnisotropy | ( | ... | ) |
Sets up directional data for anisotropic regularization.
This routine provides a two-dimensional normalized vector field 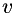 , which is normal to edges in the given data, found as the eigenvector of the structure tensor belonging to the largest eigenvalue.
, which is normal to edges in the given data, found as the eigenvector of the structure tensor belonging to the largest eigenvalue. 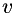 is encoded by a scalar field
is encoded by a scalar field 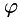 of angles, i.e.
of angles, i.e. 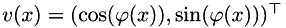 .
.
In addition, two scalar fields 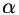 and
and 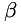 are generated from scalar parameters
are generated from scalar parameters 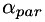 and
and 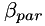 , such that
, such that
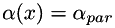 at edges, at edges, |
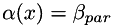 in homogeneous regions, in homogeneous regions, |
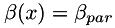 . . |
Declarations:
Output:
| Three scalar fields phi, alpha and beta. |
Input:
| data: | two-dimensional scalar field. |
| alpha_par,beta_par: | two positive values for setting up the scalar fields alpha and beta |
| sigma_par: | non-negative parameter for presmoothing the data. |
| rho_par: | non-negative parameter for presmoothing the structure tensor. |
| K_par: | positive edge sensitivity parameter. |
(see anisotropicTotalVariationFilter() and secondOrderTotalVariationFilter() for usage in an application).
- Examples:
- total_variation.cxx.
| void vigra::anisotropicTotalVariationFilter | ( | ... | ) |
Performs Anisotropic Total Variation Regularization.
The algorithm minimizes
![\[ \min_u \int_\Omega \frac{1}{2} (u-f)^2 + \sqrt{\nabla u^\top A \nabla u}\;dx\qquad\qquad(2) \]](form_109.png)
where 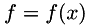 are the noisy data,
are the noisy data, 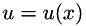 are the smoothed data,
are the smoothed data, 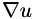 is the image gradient in the sense of Total Variation and
is the image gradient in the sense of Total Variation and 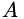 is a locally varying symmetric, positive definite 2x2 matrix.
is a locally varying symmetric, positive definite 2x2 matrix.
Matrix 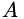 is described by providing for each data point a normalized eigenvector (via angle
is described by providing for each data point a normalized eigenvector (via angle 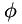 ) and two eigenvalues
) and two eigenvalues 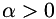 and
and 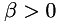 .
.
getAnisotropy() can be use to set up such data 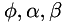 by providing a vector field normal to edges.
by providing a vector field normal to edges.
Declarations:
anisotropicTotalVariationFilter() implements a primal-dual algorithm to solve (2).
Input:
| data: | input data to be filtered. |
| steps: | iteration steps. |
| weight : | a point-wise weight ( 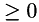 ) for the data term. ) for the data term. |
| phi,alpha and beta : | describe matrix 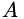 , see above. , see above. |
Output:
| out : | contains filtered data. |
Usage:
E.g. with a solution-dependent adaptivity cf. [1], by updating the matrix 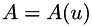 in an outer loop:
in an outer loop:
#include <vigra/tv_filter.hxx>
[1] Frank Lenzen, Florian Becker, Jan Lellmann, Stefania Petra and Christoph Schnörr, A Class of Quasi-Variational Inequalities for Adaptive Image Denoising and Decomposition, Computational Optimization and Applications, Springer, 2012.
- Examples:
- total_variation.cxx.
| void vigra::secondOrderTotalVariationFilter | ( | ... | ) |
Performs Anisotropic Total Variation Regularization.
The algorithm minimizes
![\[ \min_u \int_\Omega \frac{1}{2} (u-f)^2 + \sqrt{\nabla u^\top A \nabla u} + \gamma |Hu|_F\;dx \qquad\qquad (3) \]](form_117.png)
where 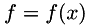 are the noisy data,
are the noisy data, 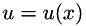 are the smoothed data,
are the smoothed data, 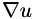 is the image gradient in the sense of Total Variation,
is the image gradient in the sense of Total Variation, 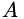 is a locally varying symmetric, positive-definite 2x2 matrix and
is a locally varying symmetric, positive-definite 2x2 matrix and 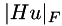 is the Frobenius norm of the Hessian of
is the Frobenius norm of the Hessian of 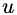 .
.
Matrix 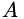 is described by providing for each data point a normalized eigenvector (via angle
is described by providing for each data point a normalized eigenvector (via angle 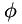 ) and two eigenvalues
) and two eigenvalues 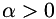 and
and 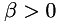 . getAnisotropy() can be use to set up such data
. getAnisotropy() can be use to set up such data 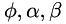 by providing a vector field normal to edges.
by providing a vector field normal to edges.
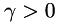 is the locally varying regularization parameter for second order.
is the locally varying regularization parameter for second order.
Declarations:
secondOrderTotalVariationFilter() implements a primal-dual algorithm to solve (3).
Input:
| data: | the input data to be filtered. |
| steps : | number of iteration steps. |
| out : | contains the filtered data. |
| weight : | point-wise weight ( 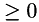 ) for the data term. ) for the data term. |
| phi,alpha,beta: | describe matrix 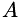 , see above. , see above. |
| xedges and yedges : | binary arrays indicating the presence of horizontal (between (x,y) and (x+1,y)) and vertical edges (between (x,y) and (x,y+1)). These data are considered in the calculation of 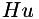 , such that finite differences across edges are artificially set to zero to avoid second order smoothing over edges. , such that finite differences across edges are artificially set to zero to avoid second order smoothing over edges. |
Usage:
E.g. with a solution-dependent adaptivity (cf.[1]), by updating the matrix 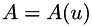 in an outer loop:
in an outer loop:
#include <vigra/tv_filter.hxx>
[1] Frank Lenzen, Florian Becker, Jan Lellmann, Stefania Petra and Christoph Schnörr, A Class of Quasi-Variational Inequalities for Adaptive Image Denoising and Decomposition, Computational Optimization and Applications, Springer, 2012.
- Examples:
- total_variation.cxx.